FairPRS: adjusting for admixed populations in polygenic risk scores using invariant risk minimization
Applying Invariant Risk Minimization (IRM) to debias ethnically-biased polygenic risk scores.
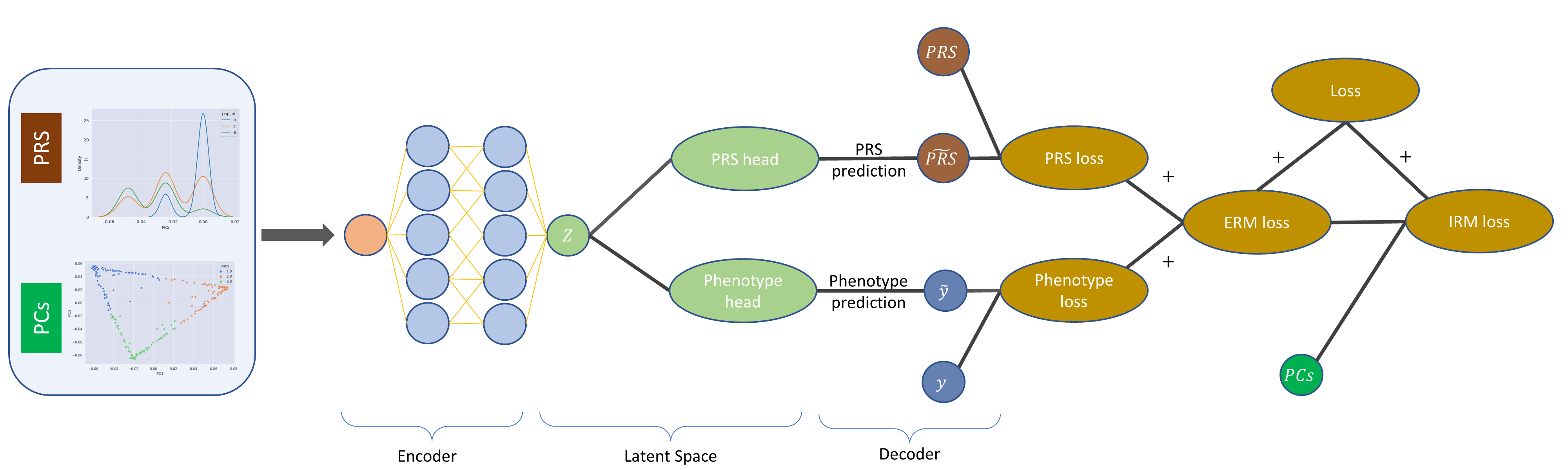
Polygenic risk scores (PRS) are increasingly used to estimate the personal risk of a trait based on genetics. However, most genomic cohorts are of European populations, with a strong under-representation of non-European groups. Given that PRS poorly transport across racial groups, this has the potential to exacerbate health disparities if used in clinical care. Hence there is a need to generate PRS that perform comparably across ethnic groups. Borrowing from recent advancements in the domain adaption field of machine learning, we propose FairPRS - an Invariant Risk Minimization (IRM) approach for estimating fair PRS or debiasing a pre-computed PRS. We test our method on both a diverse set of synthetic data and real data from the UK Biobank. We show our method can create ancestry-invariant PRS distributions that are both racially unbiased and largely improve phenotype prediction. We hope that FairPRS will contribute to a fairer characterization of patients by genetics rather than by race.
Citation
@inproceedings{reyes2023fairprs,
title={FairPRS: adjusting for admixed populations in polygenic risk scores using invariant risk minimization},
author={Reyes, Diego Machado and Bose, Aritra and Karavani, Ehud and Parida, Laxmi},
booktitle={Pacific Symposium on Biocomputing},
year={2023}
}