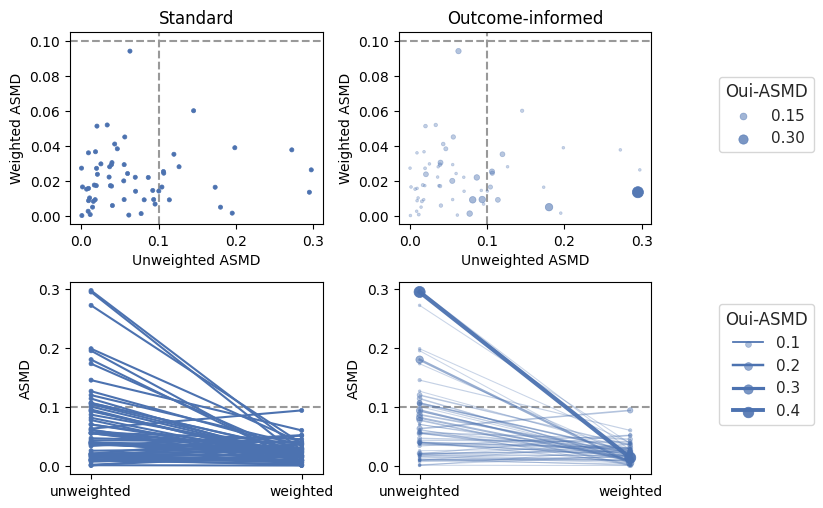
Outcome-informed balance plots
import numpy as npimport pandas as pdimport seaborn as snsimport seaborn.objects as soimport matplotlib as mplimport statsmodels.api as smimport statsmodels.formula.api as smffrom scipy.special import expitfrom sklearn.linear_model import LogisticRegression, LogisticRegressionCVfrom sklearn.linear_model import LinearRegressionfrom sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressorfrom sklearn.inspection import permutation_importancefrom sklearn.metrics import r2_score, mean_absolute_error, mean_squared_errorfrom causallib.estimation import IPW
def generate_data(n= 1000 , seed= 0 ):= 60 = np.random.default_rng(seed)= rng.normal(0 , 0.5 , size= (n, d))= np.concatenate((0 , 0.01 , size= d// 4 ),0 , 0.5 , size= d// 4 ),0 , 0.01 , size= d// 4 ),0 , 0.5 , size= d// 4 ),= X @ a_beta= expit(a_logit)= rng.binomial(1 , a_prop)= np.concatenate((0 , 0.01 , size= d// 2 ),- 2 , 0.5 , size= d// 2 ),= 1 = X @ y_beta + a * effect + rng.normal(0 , 1 , size= n)= pd.DataFrame(X, columns= [f"x { i:02d} " for i in range (X.shape[1 ])])= pd.Series(a, name= "a" )= pd.Series(y, name= "y" )return X, a, y
= generate_data()
0
0.062865
-0.066052
0.320211
0.052450
-0.267835
0.180798
0.652000
0.473540
-0.351868
-0.632711
...
-0.002227
0.328237
-0.644181
0.197561
0.214932
0.348021
-0.592059
-0.330851
0
-6.701799
1
-0.218218
-0.584901
0.869684
-0.247955
0.164485
-0.129286
0.791736
0.660180
0.316676
-1.101755
...
-0.037851
0.101057
0.347086
-0.379185
0.710491
0.363047
0.421866
0.582432
0
-2.816572
2
0.393794
0.422039
0.037797
-0.713387
-0.067523
-0.384757
-0.711371
0.129226
-0.284275
-0.514902
...
0.458327
0.185473
0.306595
-0.076096
-0.736944
0.514427
-0.967480
-0.119968
1
6.779806
3
-0.102261
-0.521430
0.306562
-0.100165
-0.218434
0.259921
-0.238290
0.694490
0.175728
-0.237166
...
0.602198
0.795350
-0.628069
-0.590841
-0.884256
-0.481927
-1.553168
-0.571139
1
6.019206
4
0.648458
-0.172836
0.427292
-0.244485
0.880334
0.099609
-0.191001
1.276212
-0.162236
-0.610612
...
0.319926
0.377184
-0.479467
0.281199
-0.145816
0.150646
-0.630480
0.416447
0
8.001258
...
...
...
...
...
...
...
...
...
...
...
...
...
...
...
...
...
...
...
...
...
...
995
0.116462
-0.830722
-0.349087
-0.073964
0.552409
-0.426036
-0.180565
0.395796
0.333374
0.634334
...
-0.533624
0.004432
0.699422
0.411842
0.089336
0.233651
-0.194621
-0.154141
0
0.727265
996
-0.162503
0.009430
0.662556
0.339446
0.935749
-0.172235
-0.291505
0.241524
0.227343
0.324185
...
0.689824
-0.811861
-0.099529
1.016822
-0.199815
0.000524
-1.422131
0.731760
0
2.401245
997
-0.073930
0.198832
-0.081691
0.164979
-0.172650
0.569894
0.120989
0.695015
0.855502
0.866506
...
0.578824
0.113620
0.035084
-0.181233
-0.260885
0.791273
0.097613
-0.702257
1
-1.878199
998
-0.086514
0.963202
-0.439070
-0.773299
0.003720
-0.676077
0.708520
0.543153
0.992313
0.430708
...
0.262656
0.536888
-0.388140
-0.087551
0.051587
-0.292679
0.170999
0.328401
1
1.401488
999
-0.330444
-0.366892
-1.220419
0.022099
0.522044
-1.146663
0.977084
-0.227534
0.036306
0.178804
...
0.386218
0.060903
-0.778868
0.843278
-0.317800
1.030502
-0.007921
-0.901038
0
-6.737274
1000 rows × 62 columns
= IPW(LogisticRegression(penalty= "none" , max_iter= 5000 ))# ipw = IPW(LogisticRegressionCV(max_iter=5000)) = ipw.compute_weights(X, a)
def calculate_asmd(X, a, w= None ):# eps = np.finfo(X.dtypes.iloc[0]).resolution # .eps = 1e-8 if w is None := pd.Series(1 , index= a.index)= a == 1 = sm.stats.DescrStatsW(X.loc[is_treated], weights= w.loc[is_treated])= sm.stats.DescrStatsW(X.loc[~ is_treated], weights= w.loc[~ is_treated])= pd.Series(x1.mean, index= X.columns)= pd.Series(x0.mean, index= X.columns)= pd.Series(x1.var, index= X.columns)= pd.Series(x0.var, index= X.columns)# smds = (x1_mean - x0_mean) / np.sqrt(x0_var + x1_var + eps) = (x1_mean - x0_mean) / ((x0_var + x1_var + eps)** 0.5 )= smds.abs ()= "asmd" return asmds
= pd.concat({"weighted" : calculate_asmd(X, a, w),"unweighted" : calculate_asmd(X, a),= ["adjustment" , "covariate" ])
adjustment covariate
weighted x00 0.015807
x01 0.029745
x02 0.036038
x03 0.029324
x04 0.017355
...
unweighted x55 0.180169
x56 0.011614
x57 0.093725
x58 0.081427
x59 0.033620
Name: asmd, Length: 120, dtype: float64
def leave_one_out_importance(estimator, X, a, y):= []for col in ["full" ] + X.columns.tolist():= X.drop(columns= col, errors= "ignore" )= curX.join(a)= estimator.predict(curXa)= {"covariate" : col,"r2" : r2_score(y, y_pred),"mse" : mean_squared_error(y, y_pred),"mae" : mean_absolute_error(y, y_pred),= pd.DataFrame(results)return resultsdef relative_explained_variation(estimator, X, a, y, metric= "mse" ):"""Harrell: https://www.fharrell.com/post/addvalue/""" = leave_one_out_importance(estimator, X, a, y)= importance.set_index("covariate" )= importance / importance.loc["full" ]= importance.drop(index= "full" )# importance = importance[metric] return importancedef decrease_in_explain_variation(estimator, X, a, y, metric= "mse" ):"""https://stackoverflow.com/q/31343563""" = leave_one_out_importance(estimator, X, a, y)= importance.set_index("covariate" )= (importance.loc["full" ]- importance) / importance.loc["full" ]= importance.drop(index= "full" )# importance = importance[metric] = importance.abs ()return importance
# i = leave_one_out_importance(LinearRegression(), X, a, y) # i = i.set_index("covariate") # i # relative_explained_variation(LinearRegression(), X, a, y) = decrease_in_explain_variation(LinearRegression(), X, a, y)
covariate
x00
5.097018e-05
0.001549
0.000339
x01
6.246515e-07
0.000019
0.000031
x02
5.112378e-05
0.001554
0.000621
x03
8.640311e-08
0.000003
0.000047
x04
2.284836e-05
0.000695
0.000077
= asmds.reset_index().merge(= "covariate" ,
0
weighted
x00
0.015807
5.097018e-05
0.001549
0.000339
1
weighted
x01
0.029745
6.246515e-07
0.000019
0.000031
2
weighted
x02
0.036038
5.112378e-05
0.001554
0.000621
3
weighted
x03
0.029324
8.640311e-08
0.000003
0.000047
4
weighted
x04
0.017355
2.284836e-05
0.000695
0.000077
...
...
...
...
...
...
...
115
unweighted
x55
0.180169
3.370652e-02
1.024676
0.426962
116
unweighted
x56
0.011614
2.034189e-02
0.618392
0.285100
117
unweighted
x57
0.093725
4.718136e-02
1.434311
0.581780
118
unweighted
x58
0.081427
5.523416e-02
1.679115
0.669075
119
unweighted
x59
0.033620
1.962519e-02
0.596605
0.274552
120 rows × 6 columns
= "mse" = plot_data.query("adjustment=='unweighted'" ).drop(columns= "adjustment" )"ouiasmd" ] = ouiasmd["asmd" ] * ouiasmd[outcome_metric]= plot_data.merge("covariate" , "ouiasmd" ]],= "covariate" ,= "left" ,= plot_data.rename(columns= {"ouiasmd" : "Oui-ASMD" })
0
weighted
x00
0.015807
5.097018e-05
0.001549
0.000339
1.466957e-05
1
weighted
x01
0.029745
6.246515e-07
0.000019
0.000031
4.835713e-07
2
weighted
x02
0.036038
5.112378e-05
0.001554
0.000621
1.453317e-05
3
weighted
x03
0.029324
8.640311e-08
0.000003
0.000047
1.459237e-07
4
weighted
x04
0.017355
2.284836e-05
0.000695
0.000077
1.351835e-05
...
...
...
...
...
...
...
...
115
unweighted
x55
0.180169
3.370652e-02
1.024676
0.426962
1.846146e-01
116
unweighted
x56
0.011614
2.034189e-02
0.618392
0.285100
7.181745e-03
117
unweighted
x57
0.093725
4.718136e-02
1.434311
0.581780
1.344308e-01
118
unweighted
x58
0.081427
5.523416e-02
1.679115
0.669075
1.367250e-01
119
unweighted
x59
0.033620
1.962519e-02
0.596605
0.274552
2.005793e-02
120 rows × 7 columns
def slope_balance(= False , = False , = "Oui-ASMD" ,= True ,= None , ax= None = so.Plot(= plot_data,= "adjustment" ,= "asmd" ,= "covariate" ,= importance_metric if opacity else None ,= importance_metric if pointsize else None ,= legend,# so.Dot(pointsize=3 if not pointsize else None), if pointsize else so.Dot(pointsize= 3 ),= importance_metric if pointsize else None ,= legend,= so.Nominal(order= ["unweighted" , "weighted" ]),= "" ,= "Absolute standardized mean difference" ,"white" )= (- 0.1 , 1.1 )if threshold is not None :0.1 , linestyle= "--" , color= "0.6" , zorder= 0 )if ax is not None := p.on(ax).plot()return p
def scatter_balance(= False , = False , = "Oui-ASMD" ,= True ,= None , ax= None = plot_data.pivot_table(# values=["asmd", "mse"] = "asmd" ,= "covariate" ,= "adjustment" "adjustment=='weighted'" ).drop(= ["adjustment" , "asmd" ]= True ,= "covariate" ,= so.Plot(= plot_data,= "unweighted" ,= "weighted" ,= importance_metric if opacity else None ,= importance_metric if pointsize else None ,# so.Dot(pointsize=3 if not pointsize else None), if pointsize else so.Dot(pointsize= 3 ),= legend,= "Unweighted ASMD" ,= "Weighted ASMD" ,"white" )if opacity:= p.scale(alpha= so.Continuous().tick(upto= 4 ))if pointsize:= p.scale(pointsize= so.Continuous().tick(upto= 4 ))if threshold is not None :0.1 , linestyle= "--" , color= "0.6" , zorder= 0 )0.1 , linestyle= "--" , color= "0.6" , zorder= 0 )if ax is not None := p.on(ax).plot()return p
= mpl.pyplot.figure(figsize= (7.5 , 5 ))= fig.subfigures(2 , 1 , hspace= 0 )= topfig.subplots(1 , 2 )= bottomfig.subplots(1 , 2 );
= 0.1 , ax= topaxes[0 ])= 0.1 , opacity= True , pointsize= True , ax= topaxes[1 ])= 0.1 , ax= bottomaxes[0 ])= 0.1 , opacity= True , pointsize= True , ax= bottomaxes[1 ])
AttributeError: 'SubFigure' object has no attribute 'savefig'
0 ].set_title("Standard" )1 ].set_title("Outcome-informed" )0 ].set_ylabel("ASMD" )1 ].set_ylabel("ASMD" )= 0.3 )= 0.3 )# fig.tight_layout()
Encoding covariate-outcome importance information (right) in scatter plots (top) and slope graphs (bottom).